Because I need to continue doing research on this topic, and it always helps me assimilate things if I write them up or otherwise explain them to others, and because I thought others here might be interested, I've decided to start a series. In each installment, I'll present one small piece of evidence for evolution. Hopefully, some of the more knowledgeable people here will chime in with related evidence.
This week: Endogenous retroviral sequencesSource: http://www.talkorigins.org/faqs/comdesc/section4.html#retroviruses
A retrovirus is a virus that makes a DNA copy of its own genome and inserts it into its host's genome. When this happens, the inserted DNA is called an endogenous retroviral sequence. Sometimes, the retrovirus inserts its DNA into the genome of a host's sperm or ovum. When this happens, all future descendants of that host will carry a copy of the retroviral DNA. The DNA becomes a sort of biological marker on that line of creature, forevermore.
It is rare for a retrovirus to make an insertion into the DNA of sperm or ovum that actually gets used in reproduction. Therefore, when two different species show the exact same endogenous retroviral sequence in the exact same place in their genome, it indicates that they descended from a single forebear.
When we examine the DNA of animals that appear related, we can tell in which order they broke off from one another by seeing which endogenous retroviral sequences they share. For example, imagine we have the following three species, with the following endogenous retroviral sequences (ERS) in the same places on their genomes:
Species A: Contains only ERS1
Species B: Contains ERS1 and ERS2
Species C: Contains ERS1, ERS2, and ERS3
This tells us two things. First, we can see that all three species are descended from a single host who was infected with ERS1 eons ago. At some point, Species B broke off as a separate line and was infected with ERS2. Later, Species C broke off from species B, carrying the first two ERS's, and was separately infected with ERS3.
Are there examples of this type of observation in real life? Yes, there are many.
The Felidae (i.e. cats) provide another example. The standard phylogenetic tree has small cats diverging later than large cats. The small cats (e.g. the jungle cat, European wildcat, African wildcat, blackfooted cat, and domestic cat) share a specific retroviral gene insertion. In contrast, all other carnivores which have been tested lack this retrogene ( Futuyma 1998 , pp. 293-294; Todaro et al. 1975 ).
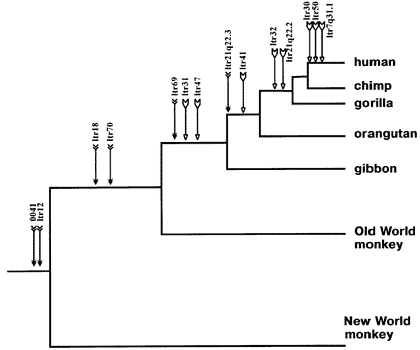
The following chart shows endogenous retroviral sequences and the branches they indicate in primates:
When scientists say that two species are related, they are not doing so based solely on the appearance or behavior of the animals. Endogenous retroviral sequences provide an empirical means of establishing the relatedness of species.
SNG
(Edited to correct spelling error)